What we do
The mammalian nervous system encompasses a vast diversity of cell-types, each defined by a unique pattern of gene expression. Our goal is to understand the gene regulatory mechanisms that specify these diverse cell-types and maintain their identity for the life of an organism. To accomplish this, we use molecular and genomic approaches to assay gene expression, chromatin structure, and the 3D folding of the genome in primary cells and tissues.
Current projects
Olfaction
Our sense of smell exhibits tremendous breadth and specificity, allowing us to identify a toxic chemical or a favorite food by scent alone. Moreover, some odorants are innately attractive or aversive, and can even induce stereotyped behaviors or physiological responses. How does the olfactory system recognize so many different odorant compounds, and what makes some odorants special, with innate valence or meaning? We try to answer these questions by combining mouse genetics with molecular and genomic analysis of olfactory neurons in vivo.
Our sense of smell exhibits tremendous breadth and specificity, allowing us to identify a toxic chemical or a favorite food by scent alone. Moreover, some odorants are innately attractive or aversive, and can even induce stereotyped behaviors or physiological responses. How does the olfactory system recognize so many different odorant compounds, and what makes some odorants special, with innate valence or meaning? We try to answer these questions by combining mouse genetics with molecular and genomic analysis of olfactory neurons in vivo.
Olfactory receptor choice
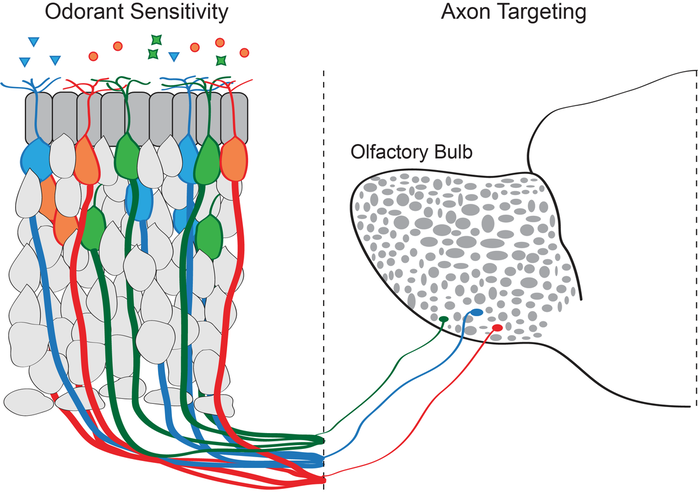
Mammalian genomes encode hundreds of olfactory receptors, allowing us to sense many diverse odorants, but each olfactory sensory neuron expresses only one type of receptor, which is determined by the stochastic and monoallelic choice of a single olfactory receptor gene for expression. This singular receptor controls olfactory neuron identity by determining the set of odorants that each olfactory neuron can recognize and by determining the set of downstream connection it will make in the olfactory bulb.
Mammalian genomes encode hundreds of olfactory receptors, allowing us to sense many diverse odorants, but each olfactory sensory neuron expresses only one type of receptor, which is determined by the stochastic and monoallelic choice of a single olfactory receptor gene for expression. This singular receptor controls olfactory neuron identity by determining the set of odorants that each olfactory neuron can recognize and by determining the set of downstream connection it will make in the olfactory bulb.
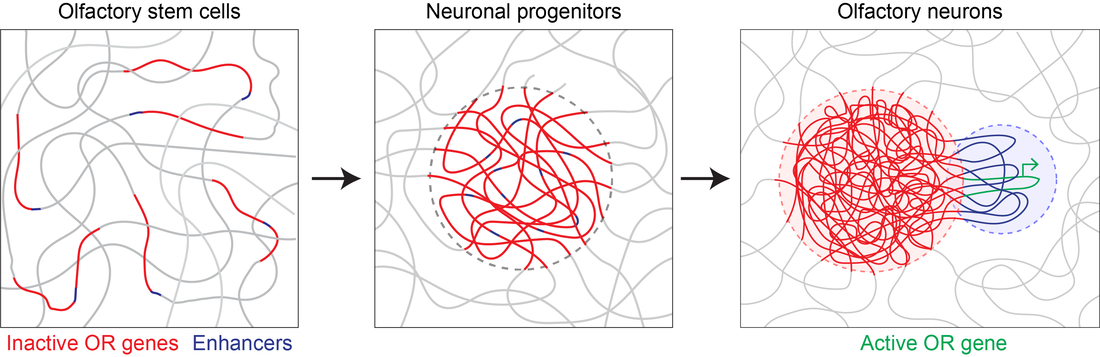
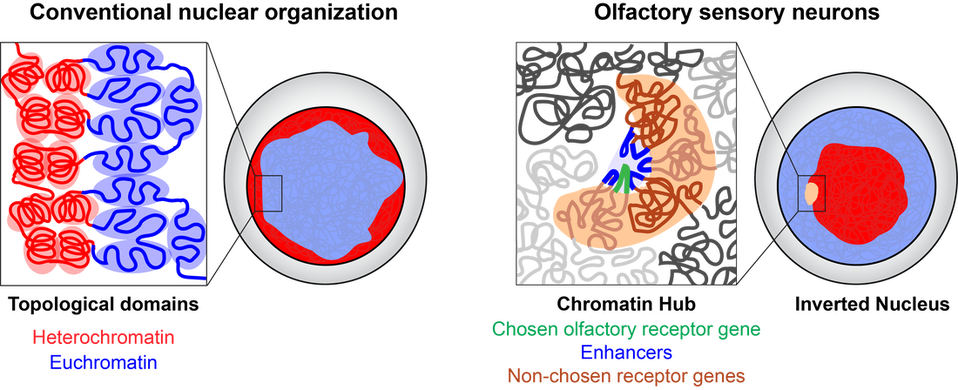
Singular olfactory receptor expression depends on remarkable changes in nuclear structure. Mice have 1300 olfactory receptor genes that are located on many different chromosomes. In most cell types these genes are dispersed throughout the nucleus, but in olfactory sensory neurons they come together in 3D space to form large heterochromatic chromatin hubs. At the same time, enhancer elements located near the olfactory receptor genes also come together to from an enhancer hub. These enhancers contact the single active olfactory receptor gene and drive its transcription.
- Developmental regulation of interchromosomal enhancer interactions - The contacts among olfactory receptor enhancers are exquisitely specific and their formation is developmentally timed to coincide with the onset of strong olfactory receptor gene expression. We are using molecular, genetic, and biochemical approaches to identify the molecular determinants that account for the specificity and developmental regulation of these contacts.
- Choice of a single olfactory receptor gene - it is unknown how multiple enhancers assemble around a single olfactory receptor gene in the first place. In order to reveal the mechanistic details of this process, we are characterizing mutant mice in which olfactory receptor expression is skewed in the favor of a few OR genes.
Vomeronasal receptor choice
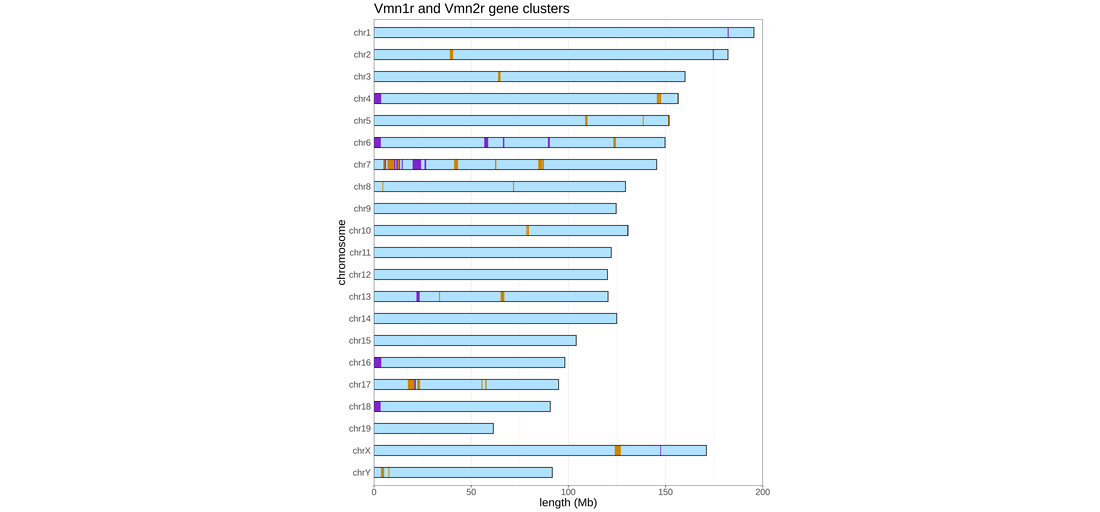
Non-volatile odorants, including many pheromones and other odorants that drive innate responses, are primarily detected by sensory neurons in the vomeronasal organ. Like olfactory receptors, vomeronasal receptors are located in large gene clusters dispersed throughout the gene, but little is known how these genes are regulated and how receptor choice is coupled to mechanisms that induce innate responses. We are using single-cell and bulk methods to analyze the role of 3d genome and chromatin structure in controlling vomeronasal receptor choice.
Non-volatile odorants, including many pheromones and other odorants that drive innate responses, are primarily detected by sensory neurons in the vomeronasal organ. Like olfactory receptors, vomeronasal receptors are located in large gene clusters dispersed throughout the gene, but little is known how these genes are regulated and how receptor choice is coupled to mechanisms that induce innate responses. We are using single-cell and bulk methods to analyze the role of 3d genome and chromatin structure in controlling vomeronasal receptor choice.
3D genome structure
Olfactory receptor choice is accompanied by a remarkable reorganization of the genome within the nucleus, involving not only the formation of olfactory receptor gene chromatin hubs, but also the pervasive repositioning of heterochromatin away from the nuclear periphery to the nuclear interior.
We are currently collaborating with Victoria Abraira's lab to examine the prevalence and function of similar rearrangements of nuclear structure in the mouse spinal cord.
Join us!
We are currently recruiting postdocs and graduate students. To learn more, email Kevin (kevin.monahan AT rutgers.edu).